
×
Eurofins Genomics can provide the synthesis of validated qPCR primers and probes for qPCR Assays recommended by the WHO and CDC.
❯❯ Order now!
×
Eurofins Genomics provides ready to use control plasmids, with one or more Coronavirus V2 gene sequences.
❯❯ Order now!
×
Eurofins Genomics gather genomics solutions for a unique market offering to support and facilitate your research for pharma, biotech and academic applications. Our scientific experts support the entire value chain of your trials though a broad range of up to date technologies and innovations.
❯❯ Read More!
×
Eurofins supports on a global scale legal authorities as well as research institutes in their fight against COVID-19.
❯❯ Learn more!
×
CE-IVD marked NovaLisa® SARS-CoV-2 (COVID-19) ELISA Kits for the qualitative detection of IgG, IgA and IgM antibodies to SARS-CoV-2 virus in human serum to aid in the diagnosis of Coronavirus.
And Immunochromatography based rapid tests that can be used outside the lab.
❯❯ Order now!
×
NovaPrime SARS-CoV-2 (COVID-19) RT-PCR Assay Kit for the qualitative detection of RNA of the novel Coronavirus in human respiratory samples.
❯❯ Order now!
×
The ViroBOAR SPIKE 1.0 RT-PCR Kit is intended for simultaneous qualitative detection of SARS-CoV-2 S gene variants in codon 501 and 570.
This leads to a discrimination of SARS-CoV-2 wildtype virus and strains B1.1.7 (UK variant) and B1.351 (South African variant).
❯❯ Order Now!
×
VIRSeek SARS-CoV-2 Assay Kits for the detection of SARS-CoV-2 in Swabs of Environmental Surfaces
❯❯ Order now!
×
Eurofins has been able to draw on its scientific experience to develop a range of SARS-CoV-2 tests in response to the Coronavirus pandemic.
❯❯ Learn more!
Coronavirus / Products & Services
qPCR PRIMERS & PROBES
Synthesis of validated qPCR primers and probes recommended by the WHO and CDC.
ORDER QPCR ASSAY

CONTROL
PLASMIDS
Ready to use COVID-19 control plasmids with one or more Coronavirus V2 gene sequences.
ORDER CONTROL PLASMIDS

TOTAL RNA SEQUENCING
High quality SARS-CoV-2 full-length genome sequencing by Next Generation Sequencing
REQUEST A QUOTE

ANTIBODY
TEST KITS
SARS-CoV-2 ELISA Kits for the qualitative detection of IgG, IgA and IgM antibodies.
ORDER ELISA KITS

MULTIPLEX
RT-PCR KIT
NovaPrime SARS-CoV-2 RT-PCR Assay Kit for the qualitative detection of RNA of COVID-19.
ORDER RT-PCR KIT

SURFACE
TEST KITS
VIRSeek SARS-CoV-2 Assay Kits for the detection of SARS-CoV-2 in swabs of environmental surfaces.
ORDER SURFACE KITS
Millions of Products / Produced under ISO & GxP
Clean, Controlled Lab for IVD and Research on SARS-CoV-2
Eurofins Genomics operates 6 facilities worldwide including operations in Europe, United States, Japan, and India.
Each production facility is producing SARS-CoV-2 material for IVD and research use under stringent quality controls. Eurofins Genomics
is accredited under ISO 13485, ISO 9001, and GMP, and it has additional CLIA and CAP accredited labs for clinical projects. In addition,
all gene and gene fragment orders are produced in completely separate facilities, isolating those sequences from DNA oligo production to
reduce the risk of cross contamination.
Lastly, Eurofins Genomics oligo production facilities have taken extra, precautionary measures to segment the downstream workflow and create
different tracks for any product that could potentially cross over, so that these tracks never cross or use the same equipment. Eurofins Genomics
is proud to be a leading supplier of testing material during the SARS-CoV-2 crisis.

Publications
- (2020) 2019 Novel Coronavirus (2019-nCoV), Wuhan, China. [Online] US Centers for Disease Control and Prevention. [Accessed 28 Jan 2020].
- (2020) 2019-Novel coronavirus (2019-nCoV) real-time rRT-PCR panel primers and probes. US Centers for Disease Control and Prevention. [Accessed 28 Jan 2020].
- (2020) Real-time RT-PCR panel for detection 2019-novel coronavirus. US Centers for Disease Control and Prevention. [Accessed 28 Jan 2020].
- (2020) Emergency use authorization. [Online] US Centers for Disease Control and Prevention. [Accessed 28 Jan 2020].
The Genomics of the Coronavirus
The coronavirus SARS-CoV-2 and resulting disease COVID-19 have reached almost every country on Earth… but compared with other infectious diseases such as the flu, there are still many unknowns about SARS-CoV-2. Scientists around the world are tirelessly researching the virus in order to understand it better and endeavor to find treatments. What is currently known about the genomics of SARS-Cov-2 and how is the information used in diagnostics?
What kind of virus is SARS-CoV-2?
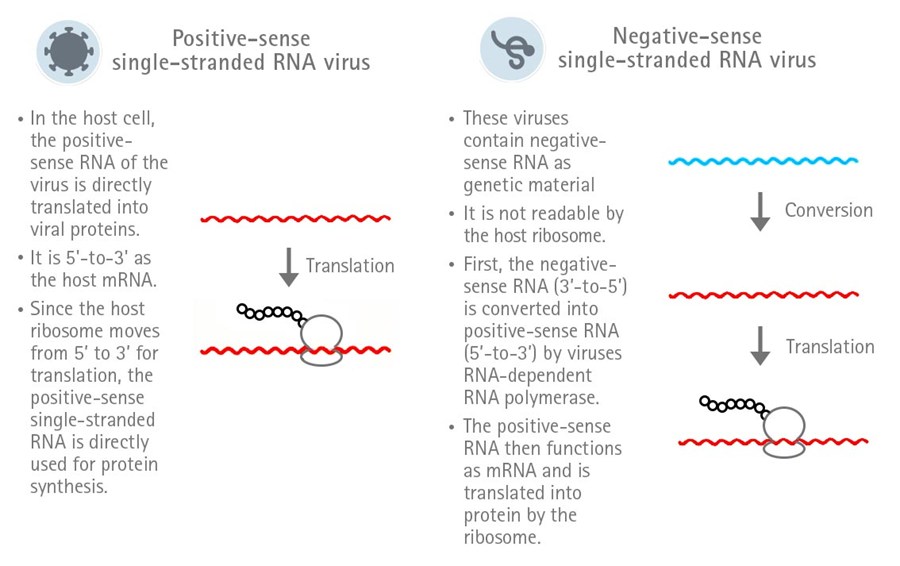
SARS-CoV-2 is a RNA virus and belongs to the family of Coronaviridae and the genus Betacoronavirus.
Its genome consists of a single-stranded positive-sense RNA. The peculiarity of single-stranded positive-sense
RNA is that it can directly function as mRNA in the host cell and, therefore, is directly translated.
In contrast, the RNA of a negative-sense single-stranded RNA virus first needs to be converted into
positive-sense RNA in order to be translated (Figure 1).
What are the genes of SARS-CoV-2?
The RNA genome of SARS-CoV-2 is approximately 30,000 nucleotides long and encodes 27 non-structural proteins and
4 structural proteins.
The 27 non-structural proteins include RNA-dependent RNA polymerase (RdRP) that was found to be highly similar
to parts of the RdRP gene of coronavirus RaTG13 in bats. Other non-structural proteins are proteases and helicases
for instance.
The 4 genes encoding structural proteins that essentially form the envelope of the virus are: 1. the spike surface
glycoprotein of the virus (S), 2. the matrix protein (M), 3. the nucleocapsid protein (N), and 3. the envelope protein (E)
(Figure 2).
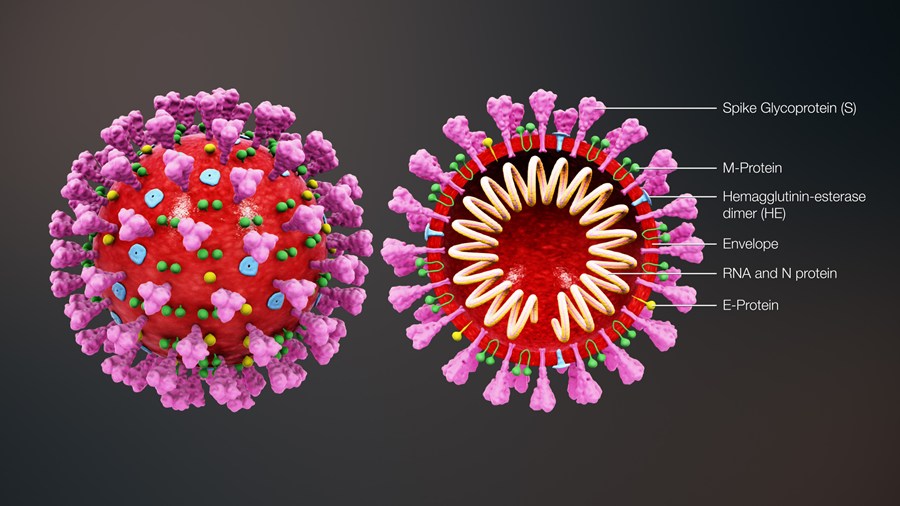
The spike proteins on the surface of the virus seem to mediate the binding of the virus to the angiotensin converting enzyme 2 (ACE2) receptor of host cells and the entry into the host cells.
The M protein seems to be involved in formation of the spherical shape of the virus.
The N protein also called nucleoprotein is associated with the single-stranded RNA and together they form the nucleocapsid.
The E protein has been found to be the smallest of the 4 structural proteins.
The genomic composition of SARS-CoV-2 has been analyzed and the sequence is publicly available at the GenBank sequence repository. As of now, 104 strains of SARS-CoV-2 have been isolated and sequenced.
How is SARS-CoV-2 identified?
Molecular techniques have been shown to be highly suitable for the identification of SARS-CoV-2 infections. Specifically real-time RT-PCR (qRT-PCR) is utilized for the analysis of respiratory samples. Here, the RNA of SARS-CoV-2 is extracted from the respiratory samples and subsequently reverse transcribed into complementary DNA (cDNA). The cDNA is then used for real-time PCR amplification with specific primers and probes that target three regions of the viral genome:
- The RdRP gene that is situated in open reading frame ORF1ab.
- The E gene
- The N gene
It has been suggested that the RdRP gene and the E genes show slightly higher analytical sensitivity than the N gene.
For identification purposes, it has been suggested to use assays that target two regions. One primer pair for the identification of a broader range of coronaviruses including SARS-CoV-2, and one primer pair that is specific for SARS-CoV-2:
- The recommendation by a research group at Charité Universitätsmedizin Berlin, Germany, that is the basis of the
WHO recommendation, is to first utilize a primer pair that targets different regions of the E gene to detect all
SARS-related viruses. In case this leads to amplification, a second analysis with a primer pair that targets
different regions of the RdRP gene is performed as confirmatory testing.
- The utilization of a primer pair that targets the N gene and one that targets the ORF1b/RNAse P gene to
confirm the results have also been proposed and are recommended by Centers for Disease Control and Prevention (CDC)
(Figure 3).
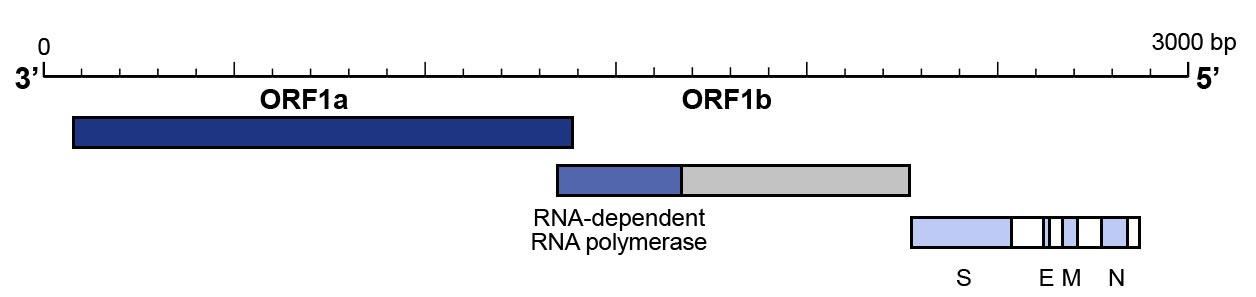
Figure 3: Genomic organization of SARS-CoV-2
qRT-PCR can be performed in a one-step or a two-step assay. The one-step assay with reverse transcription and PCR amplification in one reaction is fast and gives reproducible results for high-throughput analysis:
- The CDC utilizes a one-step qRT-PCR assay to detect a SARS-CoV-2 in samples. Here, primers and a fluorophore-quencher
probe Black Hole Quencher-1 (BHQ1) and fluorescein amidite (FAM) are utilized. During amplification the
fluorophore-quencher probe is cleaved, which results in the emission of a fluorescent signal.
- Charité Universitätsmedizin Berlin, Germany reported to also utilise a one-step qRT-PCR assay.
Here, the probes for the assay contained Blackberry Quencher (BBQ) and 6-carboxyfluorescein (6-FAM).
The two-step assay, where reverse transcription and PCR amplification are performed in different tubes, is generally more sensitive but requires more time.
The utilization of qRT-PCR assays requires positive controls for valid and reliable analyses.
What are the positive controls for SARS-CoV-2 identification by qRT-PCR?
A range of validated positive controls as plasmids that include all genes of SARS-CoV-2 as recommended
by the CDC and WHO for example is also provided: Control Plasmids for Coronavirus Research.
References
Andersen, K.G., Rambaut, A., Lipkin, W.I., Holmes, E.C., Garry, R.F. (2020) The proximal origin of SARS-CoV-2.
Nat Med. 26:450-2.
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML,
Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M,
Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. (2020) Detection of 2019 novel coronavirus (2019-nCoV) by
real-time RT-PCR. Euro Surveill. 25(3):2000045.
Phan, T. (2020) Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 81:104260
Udugama, B., Kadhiresan, P., Kozlowski, H.N., Malekjahani, A., Osborne, Li, M., V.Y. C., Chen, H., Mubareka, S., Gubbay, J.B., Chan, W.C.W. (2020) Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano [Online ahead of print].